Highly Pathogenic Avian Influenza A(H5N1) Virus Infection in a Child with No Known Exposure — San Francisco, California, December 2024–January 2025
Weekly / September 4, 2025 / 74(33);522–527
Farrell A. Tobolowsky, DO1; Eric Morris, MPH1; Lina Castro, MPH1; Tina Schaff1; Monica Jacinto1; Joseph P. Clement, MS1; Min Z. Levine, PhD2; Julia C. Frederick, PhD2; Feng Liu, PhD2; Crystal Holiday, PhD2; Marie K. Kirby, PhD2; C. Todd Davis, PhD2; Krista Kniss, MPH2; Sonja J. Olsen, PhD2; Rahil Ryder, MS3; Debra A. Wadford, PhD3; Godfred Masinde, PhD1; George Han, MD1; A. Danielle Iuliano, PhD2; Seema Jain, MD1 (View author affiliations)
View suggested citationSummary
What is already known about this topic?
As of January 1, 2025, 37 human cases of highly pathogenic avian influenza (HPAI) A(H5N1) had been detected in California, none of which occurred in San Francisco.
What is added by this report?
On January 9, 2025, a case of HPAI A(H5N1) infection was identified in a school-aged child in San Francisco through enhanced surveillance (influenza A virus subtyping of a sample of specimens weekly). No source of exposure was identified, and investigations found no laboratory evidence of human-to-human transmission among close contacts.
What are the implications for public health practice?
Enhanced surveillance and timely subtyping of a subset of influenza A-positive specimens, including specimens from persons without known A(H5N1) exposure, are important to detect avian influenza A virus infections. Public health investigations are critical to monitoring for human-to-human transmission.
Altmetric:
Abstract
In response to a highly pathogenic avian influenza (HPAI) A(H5N1) outbreak in U.S. dairy cows detected in March 2024, with subsequent identification of human cases, the San Francisco Department of Public Health instituted enhanced influenza surveillance (influenza A virus subtyping of a sample of specimens weekly) in June 2024. As of January 1, 2025, 37 human cases of influenza A(H5N1) had been detected in California, none of which occurred in San Francisco. On January 9, 2025, enhanced surveillance detected a human influenza A(H5N1) virus genotype B3.13 infection in a school-aged child in San Francisco with mild illness. Case investigation and contact tracing were conducted to ascertain exposures and detect possible human-to-human transmission. Activities comprised a household visit that included an environmental assessment, close contact interviews and surveys, and molecular and serologic testing. Sixty-seven close contacts (household, school, and health care) were identified. Upper respiratory tract specimens collected from seven asymptomatic household contacts and four symptomatic school contacts all tested negative for influenza virus by real-time reverse transcription-polymerase chain reaction (rRT-PCR). Although antibodies against influenza A(H5N1) were detected in the index patient, serologic testing of a convenience sample of nine close contacts identified no detectable A(H5)-specific antibodies. Despite an extensive investigation, the infection source remains unknown; no human-to-human transmission was identified among close contacts by rRT-PCR and serologic testing. Continued enhanced surveillance and timely subtyping of a subset of influenza A-positive specimens are essential components of a comprehensive strategy to detect human novel influenza A virus infections, including among persons without known exposures to A(H5N1) viruses.
Introduction
An outbreak of highly pathogenic avian influenza (HPAI) A(H5N1) virus in Texas dairy cows was detected in March 2024; the first associated human A(H5N1) case was identified in Texas in April 2024 (1,2). By January 1, 2025, a total of 66 human A(H5N1) cases had been detected nationwide, including 37 (56%) in California that were associated with the 2024–2025 dairy cow outbreak (3). Although historically, severity of A (H5N1) cases varied widely, recent U.S. cases have been associated with mild illnesses (4). No human-to-human transmission has been documented in the United States (CDC Report on Missouri H5N1 Serology Testing).
In response to the outbreak, the San Francisco Department of Public Health (SFDPH) strengthened influenza surveillance in June 2024 by performing additional influenza A virus subtyping, including A(H5) testing at the SFDPH Public Health Laboratory (PHL) (5,6). This additional subtyping was recommended for 1) specimens from patients with epidemiologic risk factors for HPAI and a clinically compatible illness, 2) influenza A virus specimens that were untypeable in the clinical laboratory, and 3) a sample of influenza A virus specimens submitted weekly from health care systems for enhanced surveillance. For the enhanced surveillance component, depending on local epidemiology, clinical laboratories submitted all or a random sample of influenza A–positive specimens from pediatric and adult outpatients, hospitalized patients, and emergency department patients, resulting in a total of 20–100 specimens submitted weekly.
On January 9, 2025, through the testing of an influenza A virus specimen submitted from a health care system for enhanced surveillance, SFDPH identified its first presumptive positive A(H5N1) virus infection in a young child with asthma but no other known health conditions. Case investigation and contact tracing were conducted to ascertain exposures and assess for possible human-to-human transmission.
Methods
Initial Public Health Notification and Case Investigation
After notification of the A(H5N1) virus detection, SFDPH immediately conducted a telephone interview with the patient's family and a medical chart review to assess the child's exposures, testing, and clinical course. Within 24 hours, SFDPH visited the household, collected nasal and oropharyngeal swabs from all adults and children in the household for real-time reverse transcription-polymerase chain reaction (rRT-PCR) testing, and completed an environmental assessment (e.g., evaluation of potential animal and food exposures).
Contact Investigation and Public Health Response
Close contacts were defined as persons with prolonged exposure to the patient at a distance of ≤6 feet, without use of recommended personal protective equipment (PPE) (CDC Interim Guidance). All close contacts were investigated.
Household contacts. All household contacts (three adults and four children) were interviewed to determine whether any had signs or symptoms clinically compatible with influenza (i.e., fever, cough, sore throat, shortness of breath, or conjunctivitis) 10 days before or after the patient’s illness. Nasal and oropharyngeal swabs were collected from all household contacts, and the three adult household contacts received serologic testing. Active symptom monitoring continued for 10 days after the index patient’s last positive test result.
School contacts. Close contacts were identified from the index patient’s school classrooms. A list of close contacts with absences during the 10 days preceding the patient’s illness onset through 10 days after the exposure period ended (defined as the last day that an A(H5N1)-positive specimen was obtained from the child) was requested. School close contacts with absences were interviewed to ascertain whether they had experienced recent illnesses, sought health care, and received any diagnostic testing, or treatment.
Health care worker contacts. Health care workers who had possible contact with the patient during any visit while the patient’s rRT-PCR test result was positive received an online survey. SFDPH requested information about patient contact, PPE use (i.e., eye protection, respiratory protection at least as protective as an N95 filtering facepiece respirator, gown, and gloves), signs and symptoms during the 10 days after exposure, and whether the workers had sought health care, or received diagnostic testing or treatment.
School and health care close contacts with influenza-compatible respiratory or constitutional symptoms at the time of interview were offered influenza testing. Household members, school contacts with recent respiratory illnesses, and health care workers were offered serologic testing for antibodies to A(H5N1) virus.
Laboratory Procedures
SFDPH PHL used influenza A rRT-PCR to test the patient’s respiratory specimens, followed by CDC’s A(H5) subtyping assay. rRT-PCR–positive patient specimens were amplified using multisegment (M)-RTPCR (7); libraries were then generated from these amplicons and sequenced on an Illumina MiSeq next-generation sequencing platform.* Virus genotype was determined by whole genome sequencing, and phylogenetic analysis was performed to compare specimens collected from the patient with previous human and animal specimens. Sera from the patient and the patient’s close contacts were tested at CDC for evidence of recent A(H5N1) virus infection using microneutralization (MN) assays and hemagglutinin inhibition (HI) assays against wild-type clade 2.3.4.4b viruses, A/Texas/37/2024 (B3.13), A/Michigan/90/2024 (B3.13), and A/Washington/240/2024 (D1.1); MN testing against seasonal influenza A(H1N1)pdm09 virus (A/Victoria/2570/2019) was used as a control (8). Seropositivity to A(H5N1) was defined as a neutralizing antibody titer ≥40 and HI antibody titer ≥40 (World Health Organization H5 Case Definitions). These activities were reviewed by CDC, deemed not research, and conducted consistent with applicable federal law and CDC policy.†
Results
Identification of the Index Patient
The index patient’s illness began on December 13, 2024. Signs and symptoms included, fever, abdominal pain, myalgias, and conjunctivitis, which lasted approximately 1 week and resulted in two health care visits (December 16 and 17). The first visit was to a local emergency department on December 16, where testing for influenza, respiratory syncytial virus, and COVID-19 was performed on a nasopharyngeal specimen. That day, an influenza A–positive rRT-PCR result was received, and the specimen was subsequently sent to SFDPH PHL for further testing as a part of enhanced surveillance. On January 9, 2025, the original specimen tested positive for A(H5) (cycle threshold [Ct] value approximately 29, with Ct values <38 considered positive) using the CDC assay at SFDPH PHL and was confirmed by CDC using the same A(H5) primer and probe set on January 14. A second specimen (oropharyngeal) collected by SFDPH on January 10 (25 days after the first specimen, when the child had been asymptomatic for 21 days), was positive for A(H5) (Ct value approximately 37); specimens collected 4 days later were negative.
Sequencing revealed clade 2.3.4.4b, genotype B3.13 viruses, closely related to B3.13 viruses detected in humans and animals in California (Figure 1). Phylogenetic analyses revealed that the sequences clustered together on an independent branch relative to other California human and dairy cattle sequences. Nucleotide and amino acid changes in the hemagglutinin (HA) and nucleoprotein (NP) genes were observed between the two sequences, consistent with viral replication, and no critical markers of mammalian adaptation (increased virulence or transmission risk) were identified.
Index Case Investigation
The index patient lived in an urban environment, did not travel, and had no reported exposure to dairy cows, cats, poultry, birds or other wild animals in the 10 days prior to the illness onset; the family had a pet dog. There were no animals at school, and the patient's family did not work in occupations that increase risk for A(H5N1) virus infection (handling, slaughtering, defeathering, butchering, culling, caring for, or milking infected animals). A member of the patient's family purchased raw poultry at a live bird market 2 weeks before the child's illness onset; the poultry was cooked and consumed the same day it was purchased.
Investigation of Close Contacts
Among 84 persons identified as possible contacts of the index patient (seven household, 53 school, and 24 health care), 67 (80%) met the close contact definition (Figure 2). No household contacts reported illness. School absences were reported for 34 (64.2%) school contacts, 26 (76.5%) of whom were interviewed (one teacher and parents of 25 children). All interviewed parents reported respiratory illnesses in their children, including seven who were symptomatic at the time of interview. The teacher had had influenza-compatible symptoms but was asymptomatic at the time of interview. Four persons were tested for one or more respiratory viruses (COVID-19, RSV, or influenza) previously while ill; all test results for influenza were negative. Among the 24 health care worker contacts from three facility visits (two urgent care, one emergency department), 11 (45.8%) completed a survey, including seven who had close contact with the patient; none reported influenza-compatible symptoms. All 11 available respiratory (oropharyngeal and nasal) specimens from close contacts (seven household and four school) were A(H5)-negative by rRT-PCR.
Serum specimens were collected from the index patient (32 days from onset to convalescent serum collection), three adult household contacts, two school contacts, and four health care contacts. Among these nine contacts, the median interval between their first exposure to the index patient and serum collection was 45 days (range = 9-47 days), and the median interval between their last exposure and serum collection was 26 days (range = 0-46 days). The patient had antibodies to all three wild-type A(H5N1) viruses, with elevated antibody titers in all assays, consistent with recent H5N1 infection: A/Texas/37/2024 (B3.13) (MN titer = 160, HI titer = 320); A/Michigan/90/2024 (B3.13) (MN titer = 320, HI titer = 226); and A/Washington/240/2024 (D1.1) (MN titer = 113, HI titer = 320). All nine close contacts' serology results were negative for all three wild-type A(H5N1) viruses.
Discussion
Although no exposure was identified, clinical presentation, molecular testing, and positive serology with elevated antibody titers confirmed HPAI A(H5N1) infection in this child. The absence of laboratory (molecular and serologic) evidence of current or recent A(H5N1) virus infection among close contacts suggests no human-to-human transmission. At least two other U.S. patients with confirmed A(H5N1) infection, including another unrelated pediatric patient in the San Francisco Bay Area, had no known exposure to A(H5N1) virus–infected domestic poultry, wild birds, dairy cows, or other infected animals (3,4).
Although no dairy processing facilities are located in San Francisco, the city is situated on a migratory bird route, and in 2024, A(H5) virus was detected in a live bird market, wild birds, and San Francisco wastewater (H5N1 bird flu detected in SF, first in California city wastewater). Although the family purchased poultry at a live bird market (the child did not accompany them to the market), the parents were confident that the child was not exposed to raw poultry, recent A(H5) testing in the market was negative, the cooked poultry consumption occurred more than 2 weeks before the child's symptom onset, and neither parent had evidence of infection, arguing against infected poultry exposure as the source. Although no wild bird exposure was reported, the child did spend time outside at school; therefore, environmental exposure is theoretically possible. As there were no clear risk factors or exposure to A(H5N1) virus, the infectious source remains unknown.
The genetic differences between the patient’s two positive specimens collected at separate time points likely reflect replication in the child during the intervening 25 days. Persistently positive influenza PCR test results have been previously reported, yet the duration of A(H5N1) viral nucleic acid detection and infection in humans is unknown and likely varies with virus and host factors (9). Although the second (oropharyngeal) swab collected from the patient was positive for A(H5N1) 4 weeks after the first, sequencing did not reveal mutations indicating mammalian adaptation.
Limitations
The findings in this report are subject to at least three limitations. First, because this case was identified through enhanced surveillance, which at the time included batch testing, there was a delay between specimen collection and the influenza A virus subtyping that led to detection of the case: as a result, the investigation occurred after the patient's illness had resolved and at the end of the 10-day monitoring period for close contacts, subjecting interviews to recall bias and limiting public health interventions such as real-time testing, isolation, antiviral treatment, and postexposure antiviral prophylaxis. Second, it was not possible to interview or collect respiratory and serum specimens from all close contacts; therefore, assessment of signs and symptoms and immunologic response was not comprehensive. Lastly, although household and health care contacts were assessed for asymptomatic infection, only symptomatic school contacts received molecular or serologic testing; thus, asymptomatic infection might have been missed.
Implications for Public Health Practice
A(H5N1) virus infections in humans without a clear animal exposure have rarely occurred in the United States (CDC Confirms H5N1 Bird Flu Infection in a Child in California) but have been documented in other countries where A(H5N1) viruses have circulated in wild birds for years. Continued enhanced surveillance and real-time subtyping of a subset of influenza A positive specimens at public health laboratories, including among persons without known risk for exposure to A(H5N1) virus, is an important part of comprehensive novel influenza surveillance strategies (Summer 2025 Influenza Surveillance).
Although the child had no known dairy cow exposure, sequencing results indicated that this case was associated with the 2024–2025 California dairy cow outbreak. The B3.13 genotype associated with this outbreak has also been detected in birds and felines (10), highlighting the continued transmissibility of the virus across susceptible species. Given the wild and domestic animal reservoirs for A(H5N1), a continued One Health approach supports surveillance of wild and domestic animal reservoirs for identification of additional animal cases and risk factors for cross-species and animal-to-human transmission.
Acknowledgments
The patient and the patient’s family; parents and children of close contacts; health care workers; Elizabeth Abaunza, Tochia Brewster Fleeton, Jeanette Chan, Felix Crespin, Ana Cuevas, Elaine Dekker, Natasha Desai, Kacy Diouf, Timur Durrani, Jacqueline Francis, Katherine Hernandez, Vivek Jain, Monica Kaitz Tilly, Mariel Lontoc, Laura Lucas Pablo, Susan Philip, Karen Luk, Brian Martin, Talibah Miller, Stella Morris, Vani Nimbal, Kathleen Quigley, Gabriel Sandoval, Vince-Ryan Santiago, Mustafa Sarray, Jeffrey Schmidt, Andrea Tenner, Allyson Villanueva, Douglas Walsh, Kin Wei Wu, Lisa Winston, San Francisco Department of Public Health; Daniel Dodson, Carol Glaser, Kathleen Harriman, Juliet Stoltey, California Department of Public Health; Carrie Reed, Timothy Uyeki, CDC
Corresponding author: Farrell A. Tobolowsky, Farrell.tobolowsky@sfdph.org.
1San Francisco Department of Public Health, San Francisco, California; 2Influenza Division, National Center for Immunization and Respiratory Diseases, CDC; 3California Department of Public Health.
All authors have completed and submitted the International Committee of Medical Journal Editors form for disclosure of potential conflicts of interest. No potential conflicts of interest were disclosed.
* Libraries were generated from M-RT-PCR amplicons and sequenced on an Illumina MiSeq using quarter reactions of Illumina DNA Prep but otherwise following manufacturer recommendations. The amplicons of the second specimen, A/California/227-1/2024, were further sequenced using enrichment capture with the Comprehensive Viral Research Panel (Twist Bioscience) following manufacturer recommendations for the Fast Hybridization methodology. The enriched and unenriched whole genome libraries were sequenced on an Illumina MiSeq (paired end 300 cycle kit).
† 45 C.F.R. part 46.102(l)(2), 21 C.F.R. part 56; 42 U.S.C. Sect. 241(d); 5 U.S.C. Sect. 552a; 44 U.S.C. Sect. 3501 et seq.
References
- Garg S, Reed C, Davis CT, et al. Outbreak of highly pathogenic avian influenza A(H5N1) viruses in U.S. dairy cattle and detection of two human cases—United States, 2024. MMWR Morb Mortal Wkly Rep 2024;73:501–5. https://doi.org/10.15585/mmwr.mm7321e1 PMID:38814843
- Uyeki TM, Milton S, Abdul Hamid C, et al. Highly pathogenic avian influenza A(H5N1) virus infection in a dairy farm worker. N Engl J Med 2024;390:2028–9. https://doi.org/10.1056/nejmc2405371 PMID:38700506
- Zhu S, Harriman K, Liu C, et al.; Los Angeles County H5 Response Team; California Department of Public Health H5 Laboratory Response Team. Human cases of highly pathogenic avian influenza A(H5N1)—California, September–December 2024. MMWR Morb Mortal Wkly Rep 2025;74:127–33. https://doi.org/10.15585/mmwr.mm7408a1 PMID:40080512
- Garg S, Reinhart K, Couture A, et al. Highly pathogenic avian influenza A (H5N1) virus infections in humans. N Engl J Med 2025;392:843–54. https://doi.org/10.1056/nejmoa2414610 PMID:39740051
- CDC; Association of Public Health Laboratories (APHL). Influenza virologic surveillance right size roadmap, 2nd edition. Atlanta, GA: US Department of Health and Human Services, CDC; 2022. https://www.aphl.org/aboutAPHL/publications/Documents/ID-Influenza-Right-Size-Roadmap-Edition2.pdf
- CDC. 2024–2025 influenza season: surveillance or novel influenza A and seasonal influenza viruses. Atlanta, GA: US Department of Health and Human Services, CDC; 2024. https://www.cdc.gov/bird-flu/php/monitoring-bird-flu/surveillance-novel-seasonal.html
- Zhou B, Donnelly ME, Scholes DT, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol 2009;83:10309–13. https://doi.org/10.1128/jvi.01109-09 PMID:19605485
- Levine MZ, Liu F, Bagdasarian N, et al. Neutralizing antibody response to influenza A(H5N1) virus in dairy farm workers, Michigan, USA. Emerg Infect Dis 2025;31:876–8. https://doi.org/10.3201/eid3104.250007 PMID:40053378
- Kitano T, Kitagawa D, Murata M, et al. Duration of PCR positivity by type of respiratory virus among children using a multiplex PCR test. J Med Virol 2024;96:e29890. https://doi.org/10.1002/jmv.29890 PMID:39188069
- Caserta LC, Frye EA, Butt SL, et al. Spillover of highly pathogenic avian influenza H5N1 virus to dairy cattle. Nature 2024;634:669–76. https://doi.org/10.1038/s41586-024-07849-4 PMID:39053575
 FIGURE 1. Phylogenetic tree* of highly pathogenic avian influenza (H5N1) genotype B3.13 whole genome sequences† — United States, March 2024–July 2025
FIGURE 1. Phylogenetic tree* of highly pathogenic avian influenza (H5N1) genotype B3.13 whole genome sequences† — United States, March 2024–July 2025
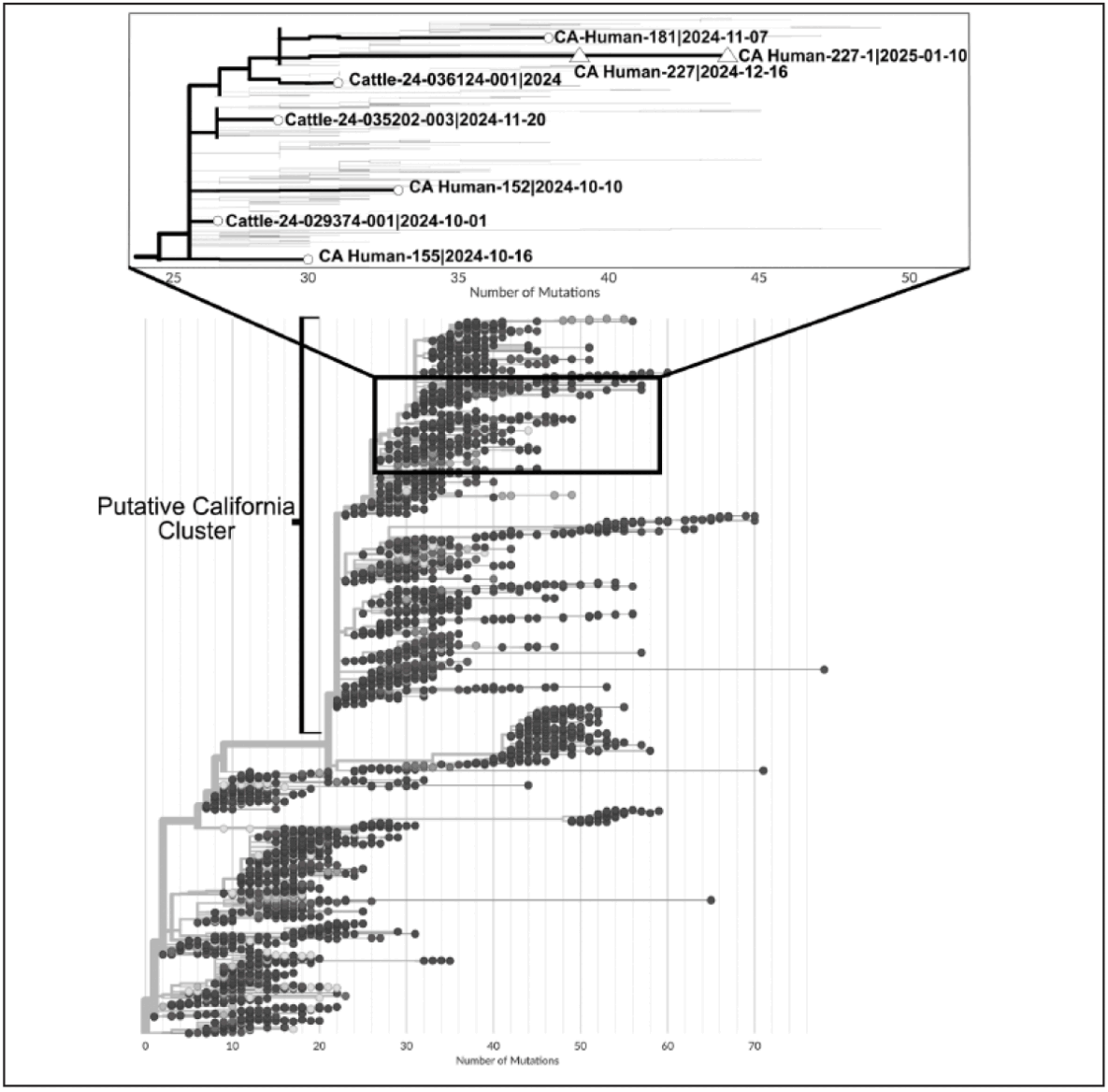
Abbreviations: HA = hemagglutinin; NP = nucleoprotein.
* Phylogenetic tree was created with the Ultrafast Sample placement on Existing tRee (UShER) tool using the pathogen option “H5N1 B3.13 (cattle) 2024 outbreak: concatenated segments.” UCSC UShER: Upload. The inset shows that the San Francisco pediatric case (CA Human-227 and CA Human 227-1) is closely related to other B3.13 cases from California. The number of mutations is relative to concatenated B3.13 cattle sequences from March 2024. Texas cattle sequences (ID: 24- 008749-007, 24-009110-018) were used for gene segments PB2, PB1, PA, NP, M, NS, and a New Mexico cattle sequence (ID: 24-010193-003) was used for gene segments HA and NA.
† (A/California/227/2024 and A/California/227-1/2024; GISAID accession: EPI_ISL_20059245; EPI_ISL_20143473). Changes of HA E34G, A156V, K325R (mature H5 HA numbering) and NP R175X (R-65%|K-35%), V270X (A-56%|V-44%), V414X (V-70%|A-30%) were identified between the first positive sample (nasal swab) and the second positive sample (oropharyngeal swab) collected 25 days apart.
 FIGURE 2. Network analysis of human A(H5N1) influenza cases and possible contacts (N = 84)* — San Francisco, California, January 2025
FIGURE 2. Network analysis of human A(H5N1) influenza cases and possible contacts (N = 84)* — San Francisco, California, January 2025

* Each node indicates a unique contact and the contact’s molecular and serologic testing status. This network diagram represents the volume of possible contacts, interview status, and their diagnostic results. The length and location of the spokes do not signify any degree of relatedness and are spaced to fit all contacts into one hub. Each hub and spoke section represents one setting for contacts, all of whom are linked to the patient. After further investigation, 67 (80%) of the 84 contacts met the close contact definition (i.e., persons with prolonged exposure to the patient at a distance of ≤6 feet, without use of recommended personal protective equipment CDC Interim Guidance).
Suggested citation for this article: Tobolowsky FA, Morris E, Castro L, et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Infection in a Child with No Known Exposure — San Francisco, California, December 2024–January 2025. MMWR Morb Mortal Wkly Rep 2025;74:522–527. DOI: http://dx.doi.org/10.15585/mmwr.mm7433a2.
MMWR and Morbidity and Mortality Weekly Report are service marks of the U.S. Department of Health and Human Services.
Use of trade names and commercial sources is for identification only and does not imply endorsement by the U.S. Department of
Health and Human Services.
References to non-CDC sites on the Internet are
provided as a service to MMWR readers and do not constitute or imply
endorsement of these organizations or their programs by CDC or the U.S.
Department of Health and Human Services. CDC is not responsible for the content
of pages found at these sites. URL addresses listed in MMWR were current as of
the date of publication.
All HTML versions of MMWR articles are generated from final proofs through an automated process. This conversion might result in character translation or format errors in the HTML version. Users are referred to the electronic PDF version (https://www.cdc.gov/mmwr) and/or the original MMWR paper copy for printable versions of official text, figures, and tables.
Questions or messages regarding errors in formatting should be addressed to mmwrq@cdc.gov.